Principal Component Analysis: A Technical Review
Principal Component Analysis
A Techical Review
Miles R. Porter
August 1, 2020
The purpose of this document is to review the concept of principal component analysis and how it relates to data analytics in general. This document will focus on the mathematical aspects of PCA, and will provide some references for implementing PCA for data analysis using python.
PCA is a technique for reducing the dimensionality of data sets. Dimensionality reduction is frequently used in data analytics to improve the performance of models (inference) as well as reduce the total amount of data needed to train a model. Consider the following motivational example of PCA.
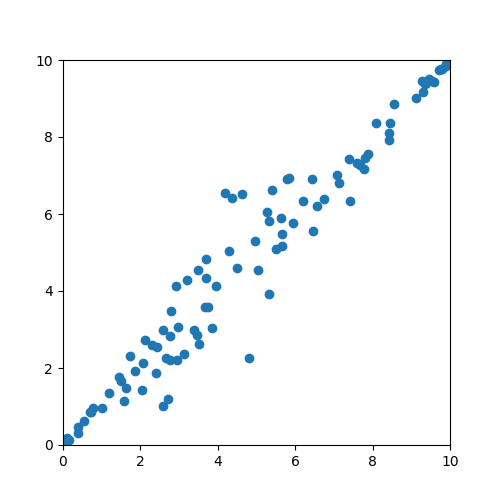
Intuitively, it seems like the data are lined up along a diagonal line. In other words, the primary dimensionality of this data is along the diagonal, and the secondary dimension of this data would be deviations from this diagonal. Consider the following image where these dimensions have been identified with red and green lines.
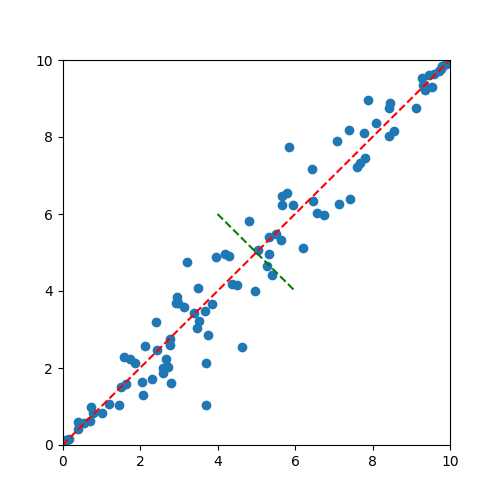
The above figure gives an intuitive feel for the goal of PCA, which is to identify the key dimensions that the data lay in. It is also important to note that in the above figure, the redline seems to capture most of the variance of the data, while the green line captures less. In this way, it is easy to see how we might consider the red line to be the primary principal component, and the green line a secondary component.
The above works well when considering data in only two dimensions, but what about higher dimensional data sets? To consider those aspects, it is important to think about the data from a more mathematical standpoint.
Moving Beyond 2 Dimensions
Consider the above example in two dimensions, but instead of thinking of the data as points on a plane think of them in matrix format, where each column of the matrix represents one of the points in the plane. Our data would then have 2 rows and 100 columns (short and very fat). The first few columns would look something like this:
[5.417.089.583.69...4.347.289.604.59...]If we consider the above matrix to be X, then we can construct a linear transformation matrix P that transforms X into a new matrix Y. This can be described mathematically as follows:
PX=YNow, the columns of X, or the xi are our original data set. Likewise, the columns of Y, or the yi are our transformed data. Our goal in PCA is to come up with a matrix P that transforms X into Y. What we are after can be considered a “change of basis”.
When we create the transformation matrix, we are going to want to accomplish a few things including finding and removing any unnecessary data. In the example above, it is clear that there are 2 dimensions that underly the data. If you imagine a third axis, it could be possible that we could reduce the data down to just two dimensions and still retain most of the valuable data. Figures 3a and 3b show a version of the above data in three dimensions. Here, clearly the Z axis is redundant as it represents very little new information that isn’t already present in the Y axis.
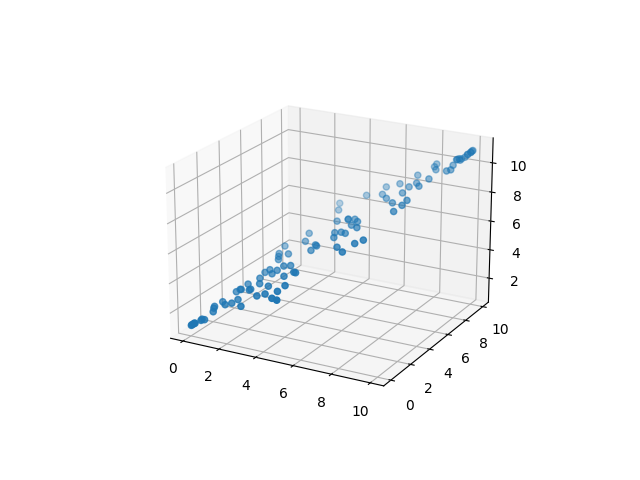

In the figure above, the red arrow indicates the principal component for this dataset. One thing to note about PCA is that once the principal components have been extracted, it is important to understand if they pertain to the new transformed space, or the original space. It is also important to keep in mind that if you transform data from a high dimensional space into a lower one, some information will be lost. It is possible to transform in reverse without loss of data only if all the principal components are used.
I have elected NOT to include code with this post for several reasons. The first of which is that I will be taking a course on computational data analytics and will likely want to reuse code I have developed for this blog post. There are, however, several good resources if you are interested in looking at specific implementations of PCA.